August 6, 2019 | Jola Glotzer
Watching CRISPR in action
Past CBC Awardee Brad Merrill, UIC, helps to resolve the CRISPR-Cas9 structure at the atomic-level before and after cutting the DNA; data recently published in Nature Structural and Molecular Biology
CRISPR-Cas9 system is a gene-editing tool that allows scientists to cut out unwanted (e.g., mutated) genomic sequences from DNA and/or add a new sequence (e.g., correct or wild-type) within a gene to change or restore its function. After Cas9 enzyme, which acts as scissors, excises a targeted DNA sequence, the cell DNA repair systems kicks in and rejoins the two separated ends of the DNA strands back together.
Prior to this work, the Cas9 structure had been solved in an inactive form, dissociated from the DNA. This publication in Nature Structural and Molecular Biology, “Cryo-EM structures reveal coordinated domain motions that govern DNA cleavage by Cas9,” adds new information to the understanding of the mechanism of Cas9 action at the exact time the enzyme is actually executing its job. Using cryo-EM, the authors observe several previously unsuspected dynamic conformational changes and rearrangements of the enzyme domains before and after it cuts the DNA. These new insights into the DNA-bound Cas9 should help improve the gene-editing properties of the CRISPR system.
The published work is a result of a collaboration between the scientists from UIC, University of British Columbia and NIH. Brad Merrill, UIC, is a co-corresponding author on the publication. He has several links to CBC: in 2014, he received a CBC Catalyst Award and participated in the CBC Tech Day as an invited speaker. Merrill was also a mentor to a CBC Scholar, Brian Shy, Class of 2013.
The CBC congratulates all scientists involved in this important discovery.
First hi-res images of active CRISPR enzyme will help improve genome editing
UIC today | by Sharon Parmet, | July 8, 2019
For the first time, scientists grappling with how to improve the efficiency of CRISPR technology — a gene-editing platform that uses an enzyme called Cas9 to precisely cut and edit specific sequences of DNA within a live cell — have captured atomic-level, three-dimensional images of the enzyme before and after cutting the DNA.
The images provide new structural information on how the enzyme works and will help researchers develop modified versions of the enzyme that can more efficiently and precisely alter targeted genes. The images and insights into Cas9 made from them are published in Nature Structural and Molecular Biology.
CRISPR is a gene-editing tool that allows scientists to cut out unwanted genes or genetic material from DNA or add a desired sequence within a gene to change its function or regulate its activity. CRISPR uses an enzyme called Cas9 that acts like scissors that cut a specific DNA sequence. Once cuts are made on either side of the DNA, the cell initiates repair systems that rejoin the two ends of the DNA strands back together.
“One of the main hurdles preventing the development of better gene-editing tools using Cas9 is that we didn’t have any images of the enzyme after the cutting DNA and did not have complete information about the changes this very important enzyme undergoes to execute the reaction,” said Miljan Simonovic, associate professor of biochemistry and molecular genetics at the University of Illinois at Chicago and a corresponding author on the paper.
Earlier structural images of Cas9 have been obtained using X-ray crystallography, but this approach has limitations. To capture various states of the enzyme in crystalline form, researchers use either inactive Cas9 (a version of the enzyme that doesn’t cleave DNA) or form the crystals under conditions that don’t support DNA cleavage. These processes can only produce images of the enzyme before it makes its cut.
“Researchers interested in either modulating Cas9 activity or engineering mutant enzymes that might work better just didn’t have complete information to begin with, so advancement was not as fast as desired,” Simonovic said. “But now we have a much clearer picture, and we even see how the major domains of the enzyme move during reaction, which may be important for further exploration.”
“It is exciting to be able to see how Cas9 actually works to cut and edit DNA strands at such a high level of detail,” said the study’s co-senior author, Sriram Subramaniam, professor of medicine at the University of British Columbia. “These images provide us with invaluable information to improve the efficiency of the gene-editing process so that we can hopefully correct disease-causing DNA mutations more quickly and precisely in the future.”
Simonovic knew a different imaging technique was needed in order to really see Cas9 at work. In the last decade, cryogenic electron microscopy, or cryo-EM, has become well known for its ability to image large molecules at high resolution under conditions that approximate their natural environments.
Simonovic and his colleagues used active Cas9 along with DNA and guide RNA. Magnesium, which is required to activate the enzyme for DNA cleavage, was added and Cas9 complexes were flash frozen and imaged using cryo-EM. The end results were snapshots of Cas9 trapped in three distinct structural arrangements while bound to nucleic acids. Most remarkably, in two of the three states, the target DNA was cleaved but the enzyme remained bound to it, allowing this final step of the biochemical reaction sequence to be observed with high resolution.
In the first state, the enzyme is captured before cutting DNA, with its major domains “assessing” if the sequence to be cut is proper. In the second, Cas9 was imaged almost immediately after the cut in the DNA is made. Practically, the enzyme’s active site hovers above the cut in the DNA strand. And, in the third state, the enzyme began its movement toward its departure from the cleaved DNA [see figure below].
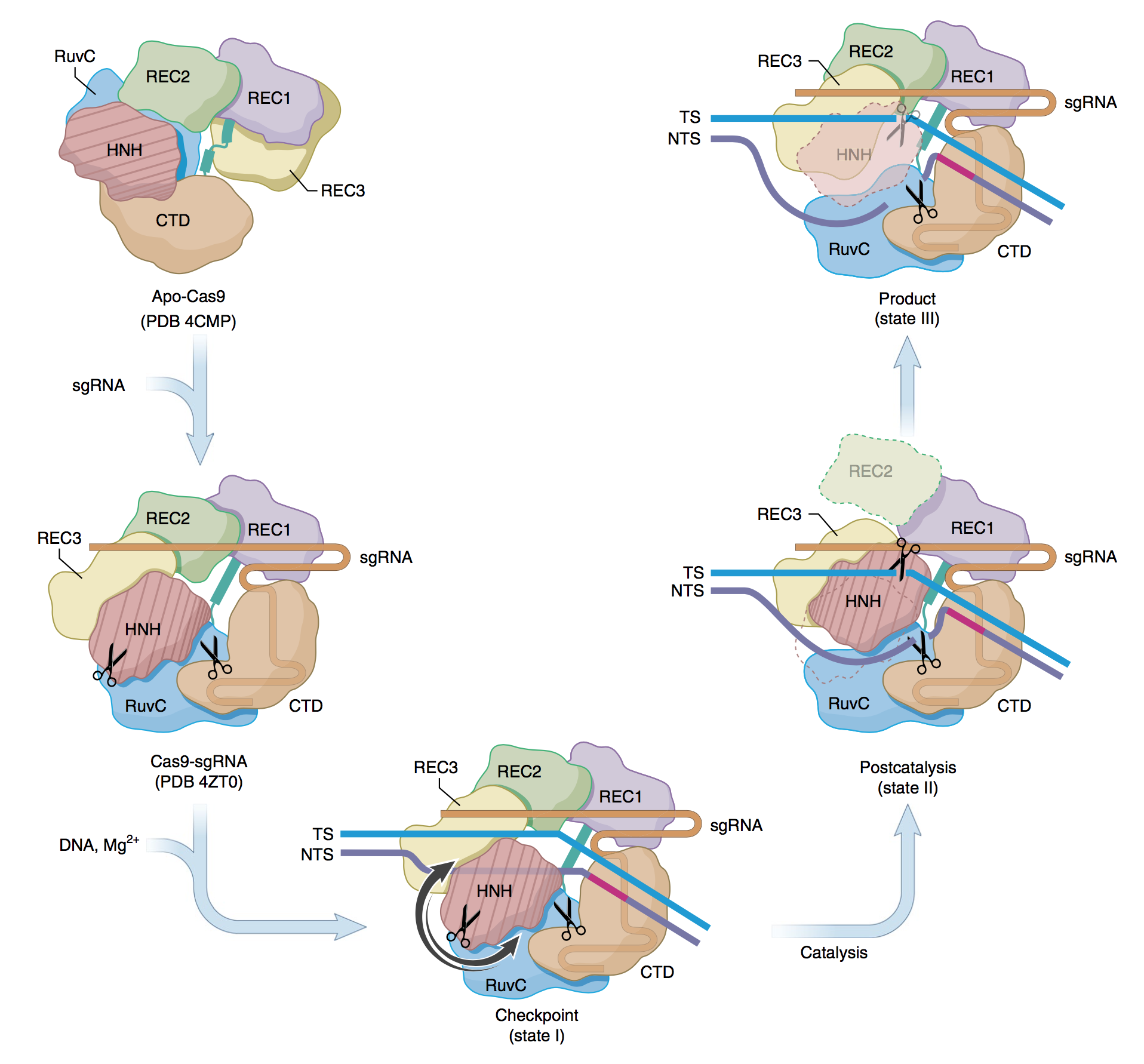
Proposed mechanism for the concerted series of domain movements involved in Cas9-mediated DNA cleavage. The binding of sgRNA to apo-Cas9 induces major domain rearrangements and formation of the binary complex. In the presence of dsDNA and Mg2+, the ‘checkpoint’ conformation (state I) is formed. The TS base pairs with the NTS and runs parallel to sgRNA, HNH adopts an inactive conformation and the distal DNA duplex is short. On activation, HNH rotates and swings (arrows indicate possible routes) towards the cleavage site in the TS. This is accompanied by REC2 disorder, ordering of REC3 loops that bind to the distal PAM and interactions between RuvC and the longer distal DNA duplex. During catalysis, the HNH and RuvC active sites (scissors) cleave the TS and NTS, respectively. After cleavage, HNH remains bound to products, whereas the RuvC active site is near the cleaved NTS. This arrangement is captured in the ‘postcatalytic’ complex (state II). In the ‘product’ complex (state III), HNH dissociates from the cleaved TS and becomes disordered and REC2 is ordered and adopts its state I conformation, while interactions between REC3, RuvC and nucleic acids persist. Apo-Cas9 and Cas9–sgRNA are based on PDB structures 4CMP and 4ZT0, respectively. (Source: www.nature.com)
“We discovered several new things about how Cas9 interacts with DNA and how it operates. One of the most interesting is how several enzyme domains move in concert and cycle between ordered and disordered states during reaction. This feature of the enzyme in which some domains are stable while others seem to be in a process of rapidly moving between different conformational states before settling down has not been seen nor predicted before,” Simonovic said. “This new information could be used to modulate how Cas9 processes targeted DNA and could help the design of better genome-editing tools.”
Bradley Merrill of UIC is a co-corresponding author on the paper. Anupama Puppala and Ryan Clarke of UIC and Xing Zhu of the University of British Columbia are co-first authors on the study. Sagar Chittori of the University of British Columbia and Alan Merk of the National Institutes of Health are co-authors on the paper.
This work was supported by the intramural research program of the National Cancer Institute, National Institutes of Health grants GM097042 and HD081534 and the UIC Center for Clinical and Translational Sciences.
Source:
Adopted (with modifications) from UIC today, by Sharon Parmet, published July 8, 2019.
Citation:
Zhu X, Clarke R, Puppala AK, Chittori S, Merk A, Merrill BJ, Simonović M, Subramaniam S. Cryo-EM structures reveal coordinated domain motions that govern DNA cleavage by Cas9. Nat Struct Mol Biol. 2019 Jul 8. [Epub ahead of print] (PubMed)
Featured CBC Community member(s):
Bradley Merrill, UIC
- CBC Catalyst Award (2014):
▸ Mapping the Regulation of Wnt Signaling with b-Cat-eleon: A Fluorescent b-catenin “timer”
PIs: Bradley Merrill (UIC) and and Fotini Gounari (UChicago) - CBC Tech Day (2014):
▸ Cutting-Edge Technologies — Driving Science Forward
Bradley Merrill (UIC) — Tech Day Speaker - CBC Scholar Award (2013-2014):
▸ Meet the Scholar: Brian Shy, UIC
PIs: Brian Shy and Bradley Merrill (mentor; UIC)
ARTICLES PUBLISHED IN THE PAST ABOUT THE FEATURED CBC COMMUNITY MEMBER(S):
July 24, 2018
▸ Problems with CRISPR
CBC past awardee and CBC Tech Day speaker, Brad Merrill, UIC, discovers a mechanism behind CRISPR gene editing imperfection
September 18, 2013
▸ Signal Transduction Factors in Breast Cancer

