June 4, 2019 | Jola Glotzer
Understanding genetic diversity
CBC Awardee, Eric Andersen, NU, explains in a recent Genome Research publication
Congratulations to Erik Andersen, NU for his contribution to a recent publication in Genome Research, titled “Long-read sequencing reveals intra-species tolerance of substantial structural variations and new subtelomere formation in C. elegans.” Using a sophisticated long-read sequencing technology, typically employed to study genomic differences between different species, the authors looked for genetic variation within the same species. To their surprise, while comparing genomes of two strains of C. elegans, Andersen and his colleagues found structural differences in 2694 genes and even formation of new subtelomeric regions. The authors concluded that mechanisms must exist allowing for such structural genomic variations to be tolerated within a species. The hypothesis that the observed variations could drive or contribute to genetic diversity within a species is also discussed.
A CBC Catalyst Award that Andersen received in 2014 for the project “Uncovering ‘Missing Heritability’ in Any Experimentally Tractable Model Organism” partially funded the current study.
Publication attributed to CBC funding*:
Kim C, Kim J, Kim S, Cook DE, Evans KS, Andersen EC, Lee J. Long-read sequencing reveals intra-species tolerance of substantial structural variations and new subtelomere formation in C. elegans. Genome Res. 2019 May 23. [Epub ahead of print] (PubMed)
ABSTRACT
Long-read sequencing technologies have contributed greatly to comparative genomics among species and can also be applied to study genomics within a species. In this study, to determine how substantial genomic changes are generated and tolerated within a species, we sequenced a C. elegans strain, CB4856, which is one of the most genetically divergent strains compared to the N2 reference strain. For this comparison, we used the Pacific Biosciences (PacBio) RSII platform (80×, N50 read length 11.8 kb) and generated de novo genome assembly to the level of pseudochromosomes containing 76 contigs (N50 contig = 2.8 Mb). We identified structural variations that affected as many as 2694 genes, most of which are at chromosome arms. Subtelomeric regions contained the most extensive genomic rearrangements, which even created new subtelomeres in some cases. The subtelomere structure of Chromosome VR implies that ancestral telomere damage was repaired by alternative lengthening of telomeres even in the presence of a functional telomerase gene and that a new subtelomere was formed by break-induced replication. Our study demonstrates that substantial genomic changes including structural variations and new subtelomeres can be tolerated within a species, and that these changes may accumulate genetic diversity within a species.
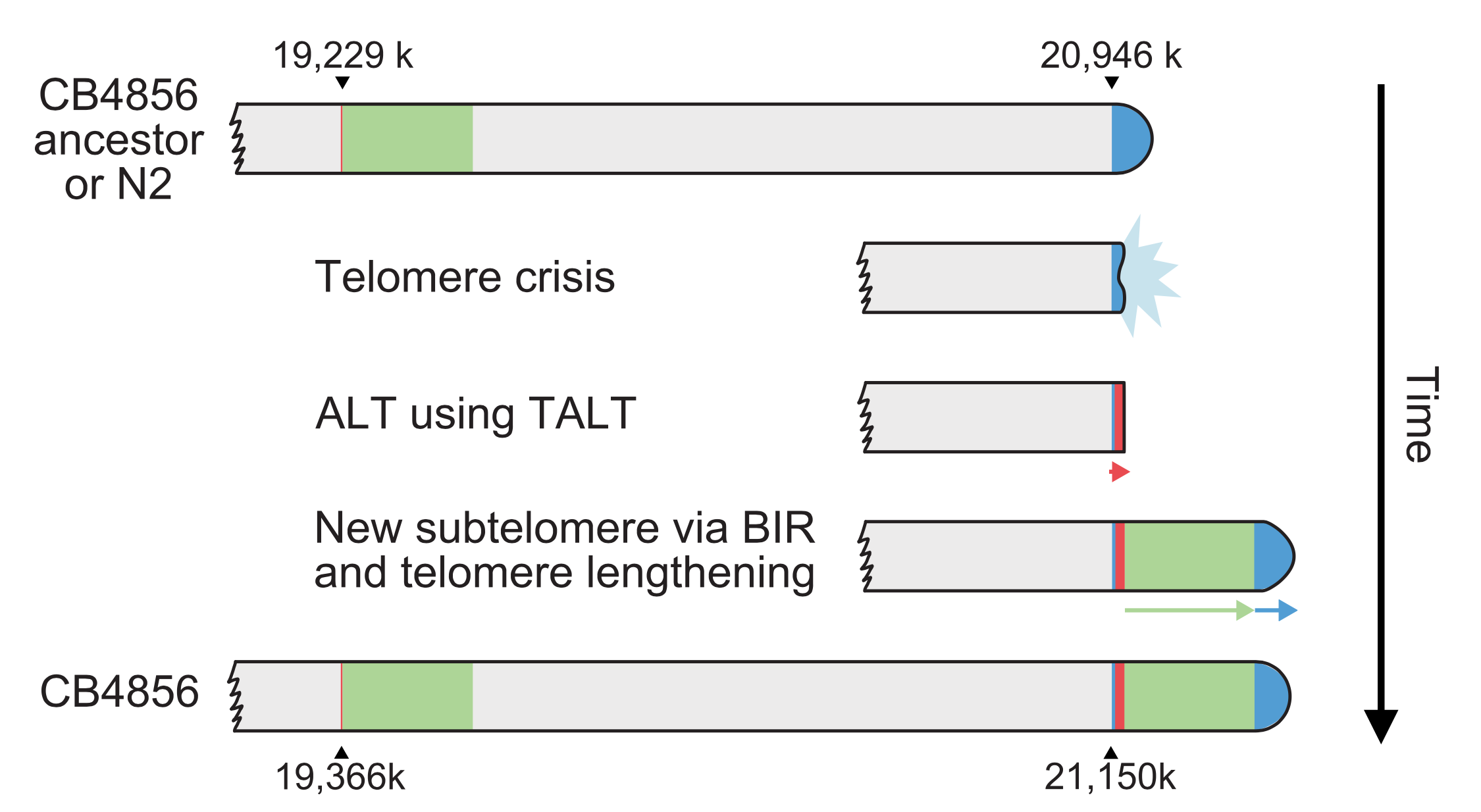
A model of Chr VR subtelomere formation in CB4856. The CB4856 ancestor underwent telomere crisis, and two sequential telo- mere-damage repair events, one using ALT and the other using BIR, formed new subtelomeres. Finally, the duplicated block end was repaired by telomerase, ending with at least 3-kb-long telomeric repeats. (Source: Genome Research)
ACKNOWLEDGMENTS
We thank B. Seo and Macrogen for initial analyses of the CB4856 genome and D. Lee for haplotype block discussion. CB4856 was kindly provided by the Caenorhabditis Genetics Center. This work was supported by the Samsung Science and Technology Foundation under Project Number SSTF-BA1501-04. C.K. was sup- ported by the BK21 program. J.K. was supported by a POSCO Science Fellowship from the POSCO TJ Park Foundation. This work was partly supported by a National Institutes of Health R01 subcontract to E.C.A. (GM107227), the Chicago Biomedical Consortium with support from the Searle Funds at the Chicago Community Trust, and an American Cancer Society Research Scholar grant to E.C.A. (127313-RSG-15-135-01-DD), along with support from the National Science Foundation Graduate Research Fellowship (DGE-1324585) to D.E.C. Additionally, we thank WormBase without which most genomic analyses would not be possible.
Featured CBC Community member(s):
Erik Andersen, NU
- *CBC Catalyst Award (2014):
▸ Uncovering ‘Missing Heritability’ in Any Experimentally Tractable Model Organism
PIs: Erik Andersen (NU) and Ilya Ruvinsky (NU; at UChicago back then)
ARTICLES PUBLISHED IN THE PAST ABOUT THE FEATURED CBC COMMUNITY MEMBER(S):
August 29, 2018
▸ 2018 NSF CAREER Award
…goes to past CBC Catalyst recipient, Erik Andersen, NU!

