June 4, 2019 | Jola Glotzer
Introducing Jump-Seq
Lulu Hu, a postdoc in Chuan He’s lab at UChicago and a recipient of a CBC Postdoctoral Research Award, publishes the results of her project in Journal of American Chemical Society
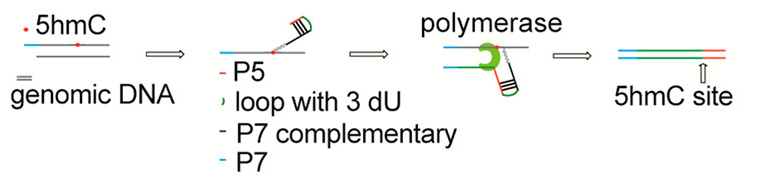
Spatio-temporal patterns of DNA de/methylation states have been implicated in many processes including embryogenesis, neurogenesis, hematopoietic development, and oncogenesis. For example, 5-hydroxymethylcytosine (5hmC) is enriched in neurons and embryonic stem cells, however it is not known what the accumulation of 5hmc does to these cell types. Hence, uncovering which genes undergo de/methylation changes in a systematic, genome-wide way, would be truly a breakthrough! Working towards this goal, Lulu Hu, along with other members of Chuan He’s lab at UChicago, developed a clever method — a Jump-seq — which allows them to capture and amplify 5hmC in genomic DNA and to identify the sequences immediately downstream of each of the 5hmC sites.
Hu is first author and He is senior author on the paper titled “Jump-Seq: Genome-Wide Capture and Amplification of 5hmC Sites,” which was published in a recent issue of in Journal of American Chemical Society. Hu’s 2016 CBC Postdoctoral Research Award, that she received for the project “Novel Sequencing Method to Profile 5mC and 5hmC in Single Cell at Base Resolution,” is acknowledged in the publication as having partially funded the study. The CBC is proud to have been able to support this work and congratulates all authors on the development of a novel and promising technology.
Publication attributed to CBC funding*:
Hu L, Liu Y, Han S, Yang L, Cui X, Gao Y, Dai Q, Lu X, Kou X, Zhao Y, Sheng W, Gao S, He X, He C. Jump-Seq: Genome-Wide Capture and Amplification of 5hmC Sites. J Am Chem Soc. 2019 May 22. [Epub ahead of print] (PubMed)
ABSTRACT
5-Hydroxymethylcytosine (5hmC) arises from the oxidation of 5-methylcytosine (5mC) by Fe2+ and 2-oxoglutarate-dependent 10–11 translocation (TET) family proteins. Substantial levels of 5hmC accumulate in many mammalian tissues, especially in neurons and embryonic stem cells, suggesting a potential active role for 5hmC in epigenetic regulation beyond being simply an intermediate of active DNA demethylation. 5mC and 5hmC undergo dynamic changes during embryogenesis, neurogenesis, hematopoietic development, and oncogenesis. While methods have been developed to map 5hmC, more efficient approaches to detect 5hmC at base resolution are still highly desirable. Herein, we present a new method, Jump-seq, to capture and amplify 5hmC in genomic DNA. The principle of this method is to label 5hmC by the 6-N3-glucose moiety and connect a hairpin DNA oligonucleotide carrying an alkyne group to the azide-modified 5hmC via Huisgen cycloaddition (click) chemistry. Primer extension starts from the hairpin motif to the modified 5hmC site and then continues to “land” on genomic DNA. 5hmC sites are inferred from genomic DNA sequences immediately spanning the 5-prime junction. This technology was validated, and its utility in 5hmC identification was confirmed.
Graphical abstract. (Source: pubs.acs.org)
ACKNOWLEDGMENTS
The authors thank Dr. Pieter Faber and the University of Chicago Genomics Facility for sequencing support. The authors also thank Dr. Kai Chen and Mr. Zhike Lu for discussion as well as Mr. Wu Tong for manuscript editing. This work was supported by the US National Institutes of Health (R01 HG006827 and P01 NS097206 to C.H.). L.H. is supported by Chicago Fellows Program, Chicago Biomedical Consortium (CBC) postdoctoral award and Leukemia & Lymphoma Society Special Fellow Award. S.G. is supported by National Key R&D Program of China (2016YFA0100400), the National Natural Science Foundation of China (31771646), and Shanghai Rising-Star Program (17QA1404200). X.H. is supported by NIH 2018R01 MH (Grant MH110531). C.H. is a Howard Hughes Medical Institute Investigator. The sequencing data reported in this paper have been deposited into the Gene Expression Omnibus (GEO) under accession number GSE127906.
Featured CBC Community member(s):
Lulu Hu and Chuan, He, UChicago
- CBC Accelerator Award (2018):
▸ A Highly Sensitive and Robust Test for Early Colorectal Cancer Diagnosis
PI: Chuan He (UChicago) - CBC Accelerator Network (CBCAN) (February 12, 2018):
▸ CBC Accelerator Network Forum
Chuan He (UChicago) — Accelerator Award program LOI presenter and award finalist - CBC Accelerator Network (CBCAN) (April 7, 2017):
▸ CBC Accelerator Network Forum
Chuan He (UChicago) — CBCAN Speaker - CBC Catalyst Award (2016):
▸ Deciphering RNA Methylation in Regulating Neuronal Functions in Health and Disease
PIs: Chuan He (UChicago) and Yongchao Ma (NU) - *CBC Postdoctoral Research Award (2016):
▸ Novel Sequencing Method to Profile 5mC and 5hmC in Single Cell at Base Resolution
PIs: Lulu Hu (postdoc) and Chuan He (UChicago) - CBC Postdoctoral Research Award (2015):
▸ Novel Sequencing Method to Profile N6-Methyldeoxyadenosine in Single-base resolution
PIs: Guan-Zheng Luo (postdoc) and Chuan He (UChicago) - CBC Postdoctoral Research Award (2014):
▸ Screening of Small Molecule Inhibitors for Nuclear RNA M6a Methyltransferase Complex
PIs: Jianzhao Liu (postdoc) and Chuan He (UChicago) - CBC Postdoctoral Research Award (2014):
▸ Pseudouridine Sequencing – A Roadmap to the New Biology of Mrna Pseudouridine Modification
PIs: Dan Dominissini (postdoc) and Chuan He (UChicago) - CBC Scholars “Loop Connections” Seminar (2013):
▸ Reversible RNA and DNA Methylation in Biological Regulation
Chuan He (UChicago) — Seminar Speaker - 10th Annual CBC Symposium (2012):
▸ Epigenomics
Chuan He (UChicago) — Symposium Speaker - CBC Catalyst Award (2011):
▸ Capturing Kinetically Labile Multiprotein Assemblies on DNA by Chemical Crosslinking
PIs: Jung-Hyun Min (UIC) and Chuan He (UChicago) - CBC Catalyst Award (2009):
▸ Virulence and latency regulation in M. tuberculosis
PIs: Chuan He (UChicago) and Scott Franzblau (UIC)
ARTICLES PUBLISHED IN THE PAST ABOUT THE FEATURED CBC COMMUNITY MEMBER(S):
May 20, 2019
▸ Nano-hmC-Seal helps identify neuroblastoma biomarkers
Four UChicago scientists with multiple ties to the CBC contribute to a recent study: Bob Grossman, Lucy Godley, Barbara Stranger and Chuan He
November 10, 2017
▸ Chuan He, University of Chicago chemist and CBC multiple-times awardee, receives the prestigious 2017 Paul Marks Prize for Cancer Research
May 22, 2012
▸ Unveiling Hidden DNA Code
March 2, 2012 (updated March 16, 2012)
▸ CBC Scholars Rock!
October 24, 2011
▸ Spotlight on October 2011 Publications by Three CBC Award Research Teams
January 14, 2015
▸ Finding Damaged DNA — Needle in the Haystack
February 17, 2016
▸ RNA Modification Discovery Suggests New Code for Control of Gene Expression

