July 12, 2019 | Jola Glotzer
Simulation-based small molecule screen
Four NU researchers and past CBC Awardees, Leonidas Platanias, Elspeth Beauchamp, Matt Clutter and Gary Schiltz, publish research funded in part by a CBC Lever Award

Leonidas Platanias, MD, PhD, the Jesse, Sara, Andrew, Abigail, Benjamin and Elizabeth Lurie Professor of Oncology and director of the NU Robert H. Lurie Comprehensive Cancer Center. (Source: NU Medicine News.)
Congratulations to a team of NU researchers, led by Leonidas Platanias, for their recent paper in the Chemical Biology & Drug Design, titled “Discovery of novel Mnk inhibitors using mutation-based induced-fit virtual high-throughput screening.” The authors describe a clever approach to screen for new inhibitors of Mnk kinase known to be involved in leukemogenesis — a process underlying the developement of acute myeloid leukemia. They use a mutation-based induced-fit (M-IFD) virtual high-throughput screening — a computer simulations of binding to a mutated kinase ATP domain by a virtual library of potential inhibitors. Indeed, the approach resulted in an identification of new molecules, further verified by other assays to have Mnk inhibitory properties. The authors conclude that, “structure-based techniques [may] support further drug discovery efforts to develop more potent and drug-like Mnk inhibitors that can potentially be used as treatments for AML and other cancers.”
The publication acknowledges CBC support as part of the work carried out at the NU Center for Molecular Innovation and Drug Discovery (CMIDD) — a facility funded by the CBC Lever Award (2009). This Award was made to Sergey Kozmin (UChicago), Karl Scheidt (NU) and Jie Liang (UIC) to support the establishment of the Chicago Tri-Institutional Center of Excellence in Chemical Methodologies & Library Development.
In addition, four NU-based co-authors on the publication have ties to the CBC: Elspeth Beauchamp, Matt Clutter and Leonidas Platanias, senior author on the paper, were co-recipients of a CBC HTS Award (2013); Gary Schiltz, corresponding author on the paper, received a CBC HTS Award (2016) and as of 2018 has been a member of the CBC Accelerator Award Review Board. CBC is grateful for Gary’s service and is proud to have supported research of all CBC community members who contributed to the current study.
Leonidas Platanias, MD, PhD, is the Jesse, Sara, Andrew, Abigail, Benjamin and Elizabeth Lurie Professor of Oncology and director of the NU Robert H. Lurie Comprehensive Cancer Center. Elspeth Beauchamp, Research Assistant Professor of Medicine (Hematology and Oncology) is a also member of the Lurie Cancer Center. Matt Clutter, PhD, Research Assistant Professor and Gary Schiltz, PhD, Research Professor, are affiliated with the Center for Molecular Innovation and Drug Discovery (CMIDD), NU Feinberg School of Medicine.
Publication attributed to CBC funding*:
Mishra RK, Clutter MR, Blyth GT, Kosciuczuk EM, Blackburn AZ, Beauchamp EM, Schiltz GE, Platanias LC. Discovery of novel Mnk inhibitors using mutation-based induced-fit virtual high-throughput screening. Chem Biol Drug Des. 2019 Jul 1. [Epub ahead of print] (PubMed)
ABSTRACT:
Mnk kinases (Mnk1 and 2) are downstream effectors of Map kinase pathways and regulate phosphorylation of eukaryotic initiation factor 4E (eIF4E). Engagement of the Mnk pathway is critical in acute myeloid leukemia (AML) leukemogenesis and Mnk inhibitors have potent antileukemic properties in vitro and in vivo, suggesting that targeting Mnk kinases may provide a novel approach for treating AML. Here, we report the development and application of a mutation-based induced-fit (M-IFD) in silico screen to identify novel Mnk inhibitors. The Mnk1 structure was modeled by temporarily mutating an amino acid that obstructs the ATP-binding site in the Mnk1 crystal structure while carrying out docking simulations of known inhibitors. The hit compounds display activity in Mnk biochemical and cellular assays, including acute myeloid leukemia progenitors. This approach will enable further rational structure-based drug design of new Mnk inhibitors and potentially novel ways of therapeutically targeting this kinase.
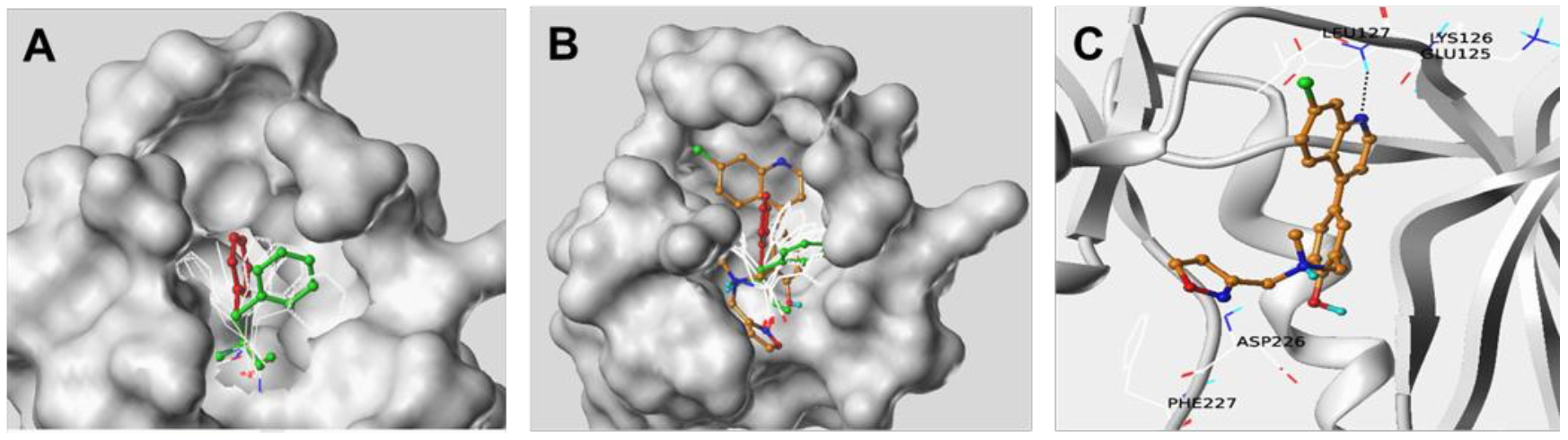
Mnk kinase structure. The average position of Phe192 as shown in Figure 1A was used for further screening of the curated small molecule kinase-focused database. The docked pose of hit compound NUCC- 54139 relative to the original Mnk1 structure and ligands used in the M-IFD is shown in Figure 1B. Key protein-ligand contacts are highlighted in Figure 1C. (Source: onlinelibrary-wiley-com)
ACKNOWLEDGMENTS:
A part of this work was performed by the Northwestern University Medicinal and Synthetic Chemistry Core (ChemCore) at the Center for Molecular Innovation and Drug Discovery (CMIDD), which is funded by the Chicago Biomedical Consortium with support from The Searle Funds at The Chicago Community Trust, and Cancer Center Support Grant P30 CA060553 from the National Cancer Institute awarded to the Robert H. Lurie Comprehensive Cancer Center.
see also:
*CBC Lever Award (2009):
▸ Chicago Tri-Institutional Center of Excellence in Chemical Methodologies & Library Development
PIs: Sergey Kozmin (UChicago), Karl Scheidt (NU) and Jie Liang (UIC)
Featured CBC Community member(s):
Elspeth Beauchamp, Matt Clutter and Leonidas Platanias, NU
- CBC HTS Award (2013):
▸ Identification of Selective mTORC2 Inhibitors for the Treatment of Cancer
PIs: Elspeth Beauchamp, Matt Clutter and Leonidas Platanias (NU)
Gary Schiltz, NU
- CBC Accelerator Review Board (ARB) (2018-present):
▸ CBC Accelerator Review Board Current Membership
Gary Schiltz, NU — Board Member - CBC HTS Award (2015):
▸ Identification of Protein Sequestration Inhibitors for Treating ALS
PIs: Gary Schiltz and Richard Silverman, NU
ARTICLES PUBLISHED IN THE PAST ABOUT THE FEATURED CBC COMMUNITY MEMBER(S):
May 14, 2019
▸ Developing sigma-2 specific probes and therapeutics
CBC Awardee Gary Schiltz, NU, contributes; CBC acknowledged for partial funding of the work recently published in ChemMedChem
May 11, 2019
▸ Will mTORC + cTORC be “it”?
Four CBC affiliates contribute to a new discovery of another TORC protein complex which promises to become a therapeutic target for some of the treatment resistant leukemias
January 4, 2018
▸ New molecule with promise to combat multiple myeloma discovered in NU center established with CBC support
October 29, 2017
▸ Embracing collaboration to combat glioblastoma
Spotlight on the Northwestern University researchers and nanotechnology
