October 1, 2019 | Jola Glotzer
Ordering proteoforms
In a recent issue of Nature Methods, CBC Senior Investigator Neil Kelleher, NU, proposes a five-level classification system for proteoform identifications
As new scientific discoveries are made and a particular area of knowledge begins to expand, it is critical to establish order in nomenclature of discovered entities, their classification into categories and systems based on identified similarities, etc. In other words, it comes down to developing a new “language” allowing scientists to communicate their findings in an organized and consistent fashion preventing confusion that could inhibit progress in the field.
In a recent Nature Methods publication, Neil Kelleher, NU, does exactly that by proposing a classification system for identifying proteoforms — a term coined only a few years ago — which is based on characteristics such as precise localization of a post-translational modification (PTM) and knowledge (or lack thereof) of the proteoform gene of origin.
Kelleher is the Walter and Mary Elizabeth Glass Professor of Chemistry, Molecular Biosciences, and Medicine at NU. He is also recognized as a CBC Senior Investigator due to his recruitment to NU in 2010 with help from the CBC Recruitment Resources Funds Award. Kelleher’s other CBC awards and community contributions are listed below the article. The CBC is proud to have played a role in Kelleher’s recruitment and to have helped support his research over the years.
Publication linked to the *CBC funding:
Smith LM, Thomas PM, Shortreed MR, Schaffer LV, Fellers RT, LeDuc RD, Tucholski T, Ge Y, Agar JN, Anderson LC, Chamot-Rooke J, Gault J, Loo JA, Paša-Tolić L, Robinson CV, Schlüter H, Tsybin YO, Vilaseca M, Vizcaíno JA, Danis PO, Kelleher NL. A five-level classification system for proteoform identifications. Nat Methods. 2019 Aug 26. [Epub ahead of print] (PubMed)
TO THE EDITOR:
The term proteoform, introduced in Nature Methods in 2013, has rapidly gained acceptance in the proteomics community. The challenge and importance of comprehensively identifying proteoforms in complex samples has been recognized, and reports have begun to appear of new platforms towards that end. However, one interesting central ambiguity has emerged, namely determining precisely what is meant by a ‘proteoform identification’. At present, the only practical approaches for establishing the exact primary structure of a proteoform employ mass spectrometry (MS), and a wide range of MS results claim proteoform identifications. This seemingly small matter has significant impact, as the ambiguity in what is meant by an ‘identification’ makes it difficult to compare results from different laboratories and approaches. This situation hinders the ability of the community to evaluate technological progress and to efficiently expand biological knowledge.
To address this issue and assist researchers in representing the ambiguity within their proteoform identifications, we propose a five-level proteoform classification system. The classification scheme covers the four types of ambiguity possible for a proteoform identification, ranging from the most subtle (that is, precise localization of a post-translational modification (PTM)), to the most dramatic (that is, ambiguity in the gene of origin).
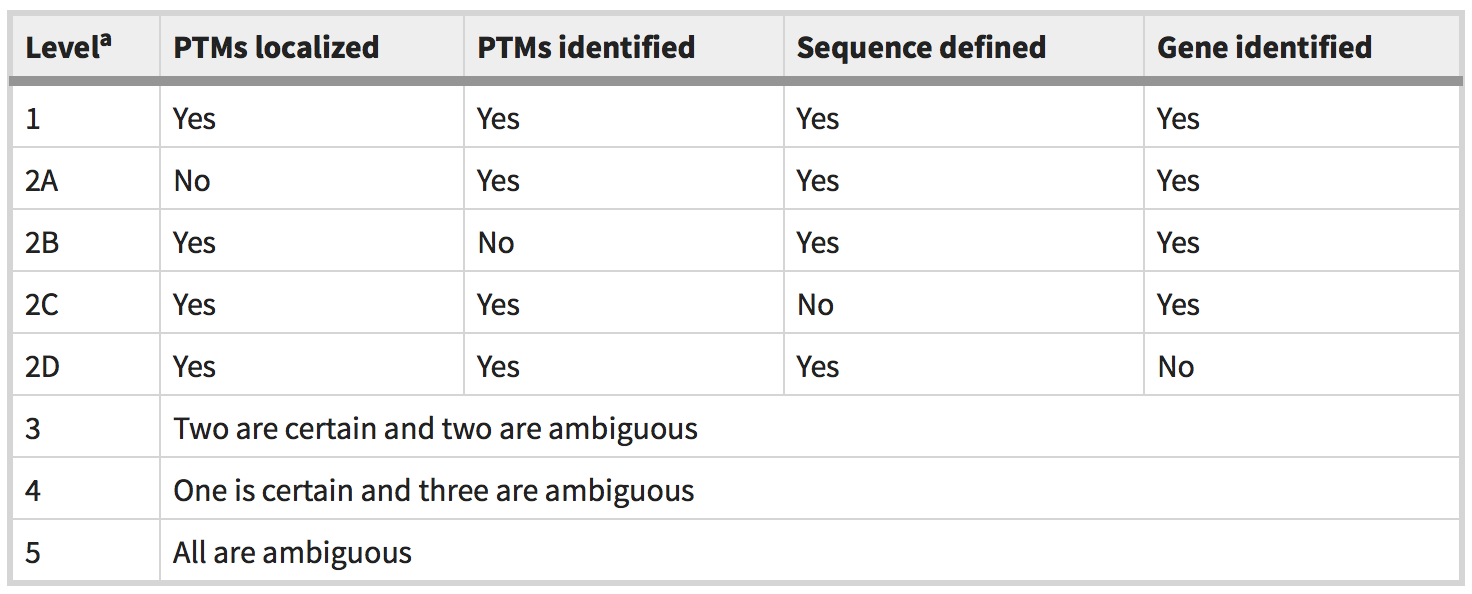
Proteoform level classification system. PTM (post-translational modification). (Source: Nat Methods)
Featured CBC Community member(s):
Neil Kelleher, NU
- CBC Exploratory Workshop (2013):
▸ The CBC Exploratory Workshop on Cellular Heterogeneity
Neil Kelleher (NU) – Workshop Organizer and Speaker - CBC Science Day (2011):
Neil Kelleher (NU) – Science Day Speaker - CBC Catalyst Award (2010):
▸ Phosphoproteomic Analysis of NADPH Oxidase Activation
PIs: Neil Kelleher (NU) and Richard Ye (UIC) - *CBC Recruitment Resources Award (2010):
▸ CBC Senior Investigator
PI: Neil Kelleher (NU) - CBC Seminar (2010):
▸ Top Down Mass Spectrometry: Has Its Decade Now Come?
Neil Kelleher (NU, UIUC then) – Seminar Speaker
Paul Thomas, NU
ARTICLES PUBLISHED IN THE PAST ABOUT THE FEATURED CBC COMMUNITY MEMBER(S):
August 6, 2019
▸ New oncogenic driver discovered
A CBC Lever Award plus a quartet of CBC awardees: Jon Oyer, Jonathan Licht and Neil Kelleher, NU, and Alex Ruthenburg, UChicago, contribute to a recent Cancer Discovery publication
July 8, 2019
▸ Defining the NSD2 interactome
Two CBC Awardees, Jonathan Licht and Neil Kelleher, identify posttranslational modification crosstalk that may play a role in carcinogenesis
July 8, 2019
▸ New insight into liver cancer treatment
CBC Awardee Rick Silverman and a CBC Senior Investigator, Neil Kelleher, NU, collaborate on developing therapeutics for hepatocellular carcinoma (HCC)
June 17, 2019
▸ Copper centers revealed with top-down mass spectrometry
Three CBC Awards contribute to today’s publication in Nature Communications!
May 13, 2019
▸ Battling epigenetic lymphomas
CBC Senior Investigator Neil Kelleher, NU, contributes to a new publication identifying a potential novel therapeutic pathway to treat the so called EZH2 dysregulated lymphomas
May 7, 2019
▸ Proteoforms explained
A recent review in Proteomics, co-authored by a CBC Senior Investigator and proteomics expert, Neil Kelleher, NU
August 9, 2018
▸ Top-down Proteomics
CBC Senior Investigator, Neil Kelleher, NU, explains the advantages of “top-down” versus “bottom-up” proteomics in early cancer detection and progression
November 13, 2017
▸ New insights into regulation of gene expression: work from the Shilatifard lab (NU) with contributions from two CBC scientists, Neil Kelleher and Jeffrey Savas.
October 5, 2017
▸ CBC Senior Investigator, Neil Kelleher, NU, deciphers molecular assembly of a gut toxin, colibactin
February 9, 2016
▸ “Clasping” Collaboration
Three CBC Scientists Join Forces to Develop Exceptional-Quality Antibodies Displaying an Unprecedented Mode of Action

